In this post I will show how to use SimpleITK to perform multi-modal segmentation on a T1 and T2 MRI dataset for better accuracy and performance. The tutorial will include input and output of MHD images, visualization tricks, as well as uni-modal and multi-modal segmentation of the datasets.
Introduction
Background
In the previous post, I introduced SimpleITK, a simplified layer/wrapper build on top of ITK, allowing for advanced image processing including but not limited to image segmentation, registration, and interpolation.
The primary strengths of SimpleITK, links and material, as well as its installation – for vanilla and alternative Python distributions – was extensively discussed in the last post entitled ‘Image Segmentation with Python and SimpleITK’. If you haven’t read that post I recommend that you do so before proceeding with this one as I will not be re-visiting these topics.
Multi-Modal Segmentation
Typically, when performing segmentations of medical image data we use a single dataset acquired through a single imaging modality, e.g., MRI, CT, PET, etc. However, different imaging modalities give us very different images of the same anatomy.
For example, a computed tomography (CT) dataset would provide a very clear depiction of bone structures in the human body. On the other hand, magnetic resonance (MR) images provide excellent contrast in soft-tissue, e.g., muscle, brain matter, etc, anatomies barely visible with CT. In addition, differently weighted images and different contrast agents depict the same anatomies with different intensities. Such a case is the gray matter which appears darker than white matter in T1-weighted images but considerably brighter than white matter in T2-weighted images.
Multi-modal segmentation essentially leverages the fact that we have multiple images of the same anatomy and allows for more precise and often faster segmentation of an anatomy. While the typical multi-modal segmentation involves combining CT and MR images (thus getting a great depiction of bones and soft tissue), I decided to keep this simple and instead show a case of gray matter segmentation based on the combination of T1 and T2 images of the same anatomy.
Dataset: The RIRE Project
Today’s dataset is taken from the Retrospective Image Registration Evaluation (RIRE) Project, which was “designed to compare retrospective CT-MR and PET-MR registration techniques used by a number of groups”.
The RIRE Project provides patient datasets acquired with different imaging modalities, e.g., MR, CT, PET, which are meant to be used in evaluation of different image registration and segmentation techniques. The datasets are distributed in a zipped MetaImage (.mhd) format with a Creative Commons Attribution 3.0 United States license which pretty much translates to “you can do whatever you want with this”. Thus, these datasets are perfect for my tutorials :).
In particular, today’s dataset is a reduced, and slightly modified, version of the ‘patient_109’ dataset which can be downloaded here. Just download my version and extract its contents alongside today’s notebook. The resulting directory structure should look something like this:
|____MultiModalSegmentation.ipynb
|____patient_109
| |____mr_T1
| | |____header.ascii
| | |____image.bin
| | |____patient_109_mr_T1.mhd
| |____mr_T2
| | |____header.ascii
| | |____image.bin
| | |____patient_109_mr_T2.mhd
|____patient_109.zip
As you can see from the above directory structure, today’s dataset comprises two MetaImage (.mhd) files with a T1 and T2 MRI datasets of a single patient. In case you didn’t know, the MHD format is a very simple format employed heavily in the distribution of medical image data. In a nutshell its just a ASCII header, with an .mhd extension, which defines basic image properties, e.g., dimensions, spacing, origin, and which is then used to read an accompanying raw binary file, typically with a .raw or .bin extension, with the actual image data. MHD files are very commonplace and inherently supported by libraries like VTK and ITK, visualization software like MayaVi, ParaView, and VisIt, as well as image processing software like 3DSlicer and MeVisLab.
Summary
In today’s post I’ll start by loading the .mhd files in the accompanying dataset and showing a few visualization tricks. Then I’ll briefly show how to smooth/denoise these images and define the seeds, operations explained in the previous post but which are still necessary for the purposes of segmentation.
Subsequently, I’ll demonstrate the uni-modal segmentation of the gray matter in each of the two images separately, i.e., T1 and T2, discussing where they fall short. Finally, I’ll show you how to combine these two image into one multi-component image which we’ll then use to perform multi-modal segmentation of the gray matter, which as we’ll see performs much better.
Multi-Modal Segmentation
Today’s IPython Notebook including the entire process can be downloaded here while you shouldn’t forget to download today’s dataset and extract it alongside the notebook.
Imports
Let’s start with the imports:
import os
import numpy
import SimpleITK
import matplotlib.pyplot as plt
%pylab inline
Once more, if you don’t have a working installation of SimpleITK, check the previous post where the topic is extensively discussed.
Helper-Functions
The following ‘helper-functions’ are defined at the beginning of today’s notebook and used throughout:
sitk_show(img, title=None, margin=0.0, dpi=40): This function usesmatplotlib.pyplotto quickly visualize a 2DSimpleITK.Imageobject under theimgparameter by first converting it to anumpy.ndarray. It was first introduced in this past post about SimpleITK.
Options
Near the beginning of today’s notebook we’ll define a few options to keep the rest of the notebook ‘clean’ and allow you to make direct changes without perusing/amending the entire notebook.
# Paths to the .mhd files
filenameT1 = "./patient_109/mr_T1/patient_109_mr_T1.mhd"
filenameT2 = "./patient_109/mr_T2/patient_109_mr_T2.mhd"
# Slice index to visualize with 'sitk_show'
idxSlice = 26
# int label to assign to the segmented gray matter
labelGrayMatter = 1
As you can see the first options, filenameT1 and filenameT2, relate to the location of the accompanying .mhd files. Again, you need to extract the contents of today’s dataset next to today’s notebook.
Unlike in the previous post, where segmentation was performed in 2D, today’s post will be performing a full 3D segmentation of the entire dataset. However, as we’ll be using the sitk_show helper-function to display the results of the segmentation, the idxSlice options gives us the index of the slice which we will be visualizing.
Lastly, labelGrayMatter is merely an integer value which will act as a label index for the gray matter in the segmentation.
Image-Data Input
We’ll start by reading the image data using SimpleITK. Since MHD images are inherently supported by ITK/SimpleIKT, reading them is as simple as this:
imgT1Original = SimpleITK.ReadImage(filenameT1)
imgT2Original = SimpleITK.ReadImage(filenameT2)
Remember:
filenameT1andfilenameT2were defined under the Options.
Next we’ll visualize these two datasets at the slice defined by idxSlice using the sitk_show helper-function. However, let’s spruce it up a bit and tile the two images using the direct call to the TileImageFilter class:
sitk_show(SimpleITK.Tile(imgT1Original[:, :, idxSlice],
imgT2Original[:, :, idxSlice],
(2, 1, 0)))
The TileImageFilter class “tiles multiple input images into a single output image using a user-specified layout”. Essentially, you pass a series of SimpleITK.Image objects, either as separate function arguments or within a list, and you get a tiled output image with the layout defined through a tuple. The result of sitk_show is shown below:

As you can see, I’ll be using direct calls to the class wrappers defined for the different filters in SimpleITK. The different between direct calls and the procedural interface offered by SimpleITK was extensively discussed in this previous post.
Image Smoothing/Denoising
As discussed in the previous post, prior to segmentation of medical image data we need to apply some smoothing/denoising to make the pixel distribution more uniform, thus facilitating region-growing algorithms.
We do so through the CurvatureFlowImageFilter class. Check the previous post for more details.
imgT1Smooth = SimpleITK.CurvatureFlow(image1=imgT1Original,
timeStep=0.125,
numberOfIterations=5)
imgT2Smooth = SimpleITK.CurvatureFlow(image1=imgT2Original,
timeStep=0.125,
numberOfIterations=5)
sitk_show(SimpleITK.Tile(imgT1Smooth[:, :, idxSlice],
imgT2Smooth[:, :, idxSlice],
(2, 1, 0)))
Note that the smoothened images now reside under imgT1Smooth and imgT2Smooth respectively. Once more, using TileImageFilter class and the sitk_show helper-function we get the next figure.

Seed Definition
Next, we need to define a series of ‘seeds’, i.e., points in the image data where we want the region-growing segmentation algorithms to start from. Again, these things were explained in the previous post. Here’s the code:
lstSeeds = [(165, 178, idxSlice),
(98, 165, idxSlice),
(205, 125, idxSlice),
(173, 205, idxSlice)]
imgSeeds = SimpleITK.Image(imgT2Smooth)
for s in lstSeeds:
imgSeeds[s] = 10000
sitk_show(imgSeeds[:, :, idxSlice])
Note that we only defined seed points on the slice defined by idxSlice so we can easily visualize them. What’s interesting here is the way we’re creating a ‘dummy’ image, i.e., imgSeeds by using the SimpleITK.Image constructor and the imgT2Smooth image. What this operation does is essentially deep-copy imgT2Smooth into imgSeeds allowing us to ruin the latter without affecting the former. We then loop through the seeds defined in lstSeeds and set those pixels to a high value so they will stand out and allow us to see them through sitk_show. The resulting image is the following:

As explained in the previous post about SimpleITK, the index order in SimpleITK is the reverse from that in NumPy. Please do read up on that part as it can be confusing and remember that the result of the visualization is based on the conversion of the SimpleITK.Image object to a numpy.ndarray one.
Auxiliary Function: Vector-Image Tiling
Before I go on with the segmentation, I want to draw your attention to the following: Those who read the previous post on SimpleITK might remember that we used the LabelOverlayImageFilter class to create overlays of the medical image and the segmented label, giving us a good idea of how (un)successful the segmentation was.
However, if we wanted to tile two overlays of the two T1 and T2 segmented images that’s when we run into trouble. The TileImageFilter class only works on single-components images, i.e., images with one value per pixel. Thus, we can’t directly tile RGB images or, in our case, images with a label-overlay.
What we can do, however, is extract the matching components from each multi-component image, tile them, and then compose the different single-component images into a multi-component one. This is exactly what the sitk_tile_vec auxiliary function we’re defining next does:
def sitk_tile_vec(lstImgs):
lstImgToCompose = []
for idxComp in range(lstImgs[0].GetNumberOfComponentsPerPixel()):
lstImgToTile = []
for img in lstImgs:
lstImgToTile.append(SimpleITK.VectorIndexSelectionCast(img, idxComp))
lstImgToCompose.append(SimpleITK.Tile(lstImgToTile, (len(lstImgs), 1, 0)))
sitk_show(SimpleITK.Compose(lstImgToCompose))
This auxiliary function takes a list of multi-component images, which in our case is going to be a list of images where we overlay the segmented label, tiles them, and uses the sitk_show helper-function to display the result. Using the GetNumberOfComponentsPerPixel method of the SimpleITK.Image class we loop through the different components in each image.
We then loop through all images in lstImgs and use the VectorIndexSelectionCastImageFilter to extract a single-component image which we append to lstImgToTile. We then tile all the single-component images and append to lstImgToCompose for the eventual composition.
Finally, we use the ComposeImageFilter, to create a single multi-component image of the tiled single-component images which we finally display with sitk_show.
Uni-Modal Segmentation
We’ll first start by applying the region-growing algorithm to each of the two images, namely imgT1Smooth and imgT2Smooth, so we can see the advantages offered by multi-modal segmentation later on.
We’re doing so using the ConfidenceConnectedImageFilter class. I suggest you take a look at the docstring for that class to see what it does but in a nutshell “this filter extracts a connected set of pixels whose pixel intensities are consistent with the pixel statistics of a seed point”. Its really all based on a confidence interval based on the multiplier parameter and the calculated standard deviation in a pixel-neighborhood. Here’s the code we’re using for the segmentation:
imgGrayMatterT1 = SimpleITK.ConfidenceConnected(image1=imgT1Smooth,
seedList=lstSeeds,
numberOfIterations=3,
multiplier=1,
replaceValue=labelGrayMatter)
imgGrayMatterT2 = SimpleITK.ConfidenceConnected(image1=imgT2Smooth,
seedList=lstSeeds,
numberOfIterations=3,
multiplier=1,
replaceValue=labelGrayMatter)
imgT1SmoothInt = SimpleITK.Cast(SimpleITK.RescaleIntensity(imgT1Smooth),
imgGrayMatterT1.GetPixelID())
imgT2SmoothInt = SimpleITK.Cast(SimpleITK.RescaleIntensity(imgT2Smooth),
imgGrayMatterT2.GetPixelID())
sitk_tile_vec([SimpleITK.LabelOverlay(imgT1SmoothInt[:,:,idxSlice],
imgGrayMatterT1[:,:,idxSlice]),
SimpleITK.LabelOverlay(imgT2SmoothInt[:,:,idxSlice],
imgGrayMatterT2[:,:,idxSlice])])
Note that much like what was done in the previous post, we create two rescaled integer versions of imgT1Smooth and imgT2Smooth named imgT1SmoothInt and imgT2SmoothInt respectively. We do so in order to create label overlays using the ‘LabelOverlayImageFilter’.
Also, note that we’re using the auxiliary function sitk_tile_vec we defined previously to tile these label overlays. The result of sitk_tile_vec is shown below.
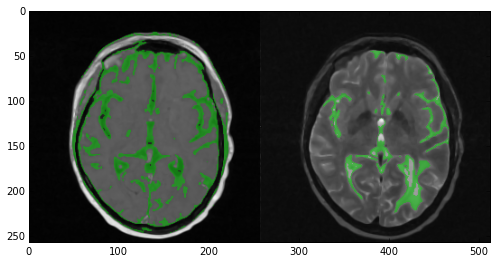
Now take a look at the results of the segmentation. In the T1 image (left) the algorithm did quite a good job covering the majority of the gray matter. However, multiple areas of the skin and fat were also included. In the case of the T2 image (right) the results were nowhere near as good. Multiple areas of gray matter were not segmented despite the large number of iterations. You can experiment with the numberOfIterations and multiplier parameters if you want but chances are you’ll end up segmenting the entire head. Alternatively you can apply more stringent criteria and up the number of seed points. However, here’s where multi-modal segmentation comes into play.
Multi-Modal Segmentation
As discussed, multi-modal segmentation gives us multiple views of the same anatomies. In our case by combining the T1 and T2 images we get two significantly different views of the gray matter and areas that were not connected in one of the images may be connected in the other. In addition, specious connections appearing in one image due to excessive noise or artifacts will most likely not appear in the other.
This allows us to perform the segmentation having more information in our arsenal and achieving the same or better results with more stringent criteria and far fewer iterations. Let’s see the code:
imgComp = SimpleITK.Compose(imgT1Smooth, imgT2Smooth)
imgGrayMatterComp = SimpleITK.VectorConfidenceConnected(image1=imgComp,
seedList=lstSeeds,
numberOfIterations=1,
multiplier=0.1,
replaceValue=labelGrayMatter)
sitk_show(SimpleITK.LabelOverlay(imgT2SmoothInt[:,:,idxSlice],
imgGrayMatterComp[:,:,idxSlice]))
Firstly, we begin by ‘combining’ the two images. In order to do that we simply use the ComposeImageFilter which we also used in the sitk_tile_vec auxiliary function. Through that filter we create a single multi-component image containing the information of both T1 and T2 images. The result of the composition is the imgComp image.
Then all we need to do is perform the segmentation. However, as you see in the above code, we don’t just use the ConfidenceConnectedImageFilter class but rather the VectorConfidenceConnectedImageFilter class. The mechanics are exactly the same with the pivotal difference being that VectorConfidenceConnectedImageFilter performs the operation on a vector image, i.e., an image with multiple components per pixel (as is our case).
Note that we’re using the same seeds but much more stringent criteria, i.e., low multiplier value, and only 1 iteration! The result can be seen below:

As you can see, the results are quite great. Not only did we get rid of the skin and fat parts that segmentation on the T1 image gave us, but we also segmented pretty much all gray matter! Of course the segmentation is not perfect as multiple areas of white matter are also segmented but given our lazy approach they’re pretty decent.
Finally, we can store the results of our segmentation as a separate .mhd file containing only the gray matter label. As we saw when loading the input .mhd files, SimpleITK makes these type of operation ridiculously easy. In order to save imgGrayMatterComp under the filename GrayMatter.mhd all we need to do is the following:
SimpleITK.WriteImage(imgGrayMatterComp, "GrayMatter.mhd")
You can download the .mhd and accompanying .raw directly from the Bitbucket repo of this blog. Now we can open this file with any of the software supporting the MHD format. In our case I opened it in ParaView and used the built-in Contour filter to get a 3D iso-surface of our segmentation which you can see in the next figure.

Links & Resources
Material
Here’s the material used in this post:
- IPython Notebook showing the presented process.
- MRI Dataset of a patient’s anatomy acquired with T1 and T2 weighting.
- Gray Matter Segmentation: the
.mhdand accompanying.rawfile containing the results of the segmentation.
See also
Check out these past posts which were used and referenced today or are relevant to this post:
- IPython Notebook & VTK
- NumPy to VTK: Converting your NumPy arrays to VTK arrays and files
- DICOM in Python: Importing medical image data into NumPy with PyDICOM and VTK
- Surface Extraction: Creating a mesh from pixel-data using Python and VTK
- Image Segmentation with Python and SimpleITK
Don’t forget: all material I’m presenting in this blog can be found under the PyScience BitBucket repository.

Pingback: Volume Rendering with Python and VTK | PyScience
Hi friend,
I am developing vtk based medical processing for cardiac centerline extractions in c#.
I am in new to this..kindly help to me…
LikeLike
Hi there,
well given that you didn’t specify what you’d like help with (and since this blog is on Python rather than C#) all I can do to help is tell you the following:
1) To my knowledge VTK doesn’t have built-in functionality for vessel-tree centerline extraction so your best bet is to:
2) Look into and use VMTK which as far as I know doesn’t wrap to C# some is written in C++ and provides Python bindings.
Take a look at the following tutorial for centerline-extraction with VMTK:
http://www.vmtk.org/tutorials/Centerlines.html
Also, the vmtk-users mailing list is rather active and the VMTK developers are very helpful:
https://groups.google.com/forum/#!forum/vmtk-users
Apart from that you’d have to be more specific with what kind of help you need and I’ll do what I can 🙂
Cheers,
Adam
LikeLike
I was trying to load other ‘.mhd’ files(downloaded from internet) but they are not loading properly ;it is throwing error as ‘RuntimeError: Exception thrown in SimpleITK ReadImage: ..\..\..\..\..\..\Build\ITK\SimpleITK-0.8.1\Code\IO\src\sitkImageReaderBase.cxx:50:
sitk::ERROR: Unable to determine ImageIO reader’
Please help
LikeLike
Dude when you give me an error like that but not the link to the images you can’t possibly expect me to be able to help right 🙂 ?. Got a link so I can try them myself? Have you tried opening the files in some software like Paraview/VisIt/3DSlicer?
LikeLike
Hello. If I wand to denoise the whole volume of a dicom file, how can I do it?
LikeLike
Just smoothen/denoise the same way shown here. The smoothing algorithms in SimpleITK aren’t limited to a single 2D slice at all. As you can see under https://pyscience.wordpress.com/2014/10/19/image-segmentation-with-python-and-simpleitk/ the SimpleITK algorithms shown were simply limited to a 2D slice to make the process faster but in your case just use the whole volume
LikeLike
Hello,
First, thank you very much for your great tutorials 🙂
But i am kinda stuck.
I have a DICOM Dataset and 3DSegmentated it like mentioned in this post: https://pyscience.wordpress.com/2014/10/19/image-segmentation-with-python-and-simpleitk/
Now i want to do Volume Rendering on this Image. I converted it with NumPy to an VTK Image, but dont know how to go on, i guess i have to label the segmentated object somehow, to get an .mhd file out of it, so that i can do volume rendering according to this post : https://pyscience.wordpress.com/2014/11/16/volume-rendering-with-python-and-vtk/
If you know any better way, it would be very nice if you can help me 🙂
LikeLike
Hey there, if you’re having trouble converting from a fancy format that only SimpleITK can work with to a proper VTK labelfield I’d suggest you use 3DSlicer and do it manually if its a one-off. I’ve been meaning to write a post on how to convert SimpleITKNumPyVTK but I haven’t found the time in forever. What format is your segmentation in?
LikeLike
I am using SITK and VTK for image segmentation. The filters and visualization functions in both toolkits are awesome. However, when using vtkDICOMImageReader, I found that the IO doesn’t include patient position and orientation data. When exporting the segmented data to other software (e.g. 3D slicer, Mimics), the spatial position of the data is incorrect. I have checked some documentations of VTK and ITK, seems there are no solution yet.
I think a coordinate transform matrix is needed to match the MRI space to VTK/ITK image space. Any idea on that?
LikeLike
Hi Jacky. Hard to say, DICOM is pretty much a free-form format and one can have any arbitrary number of key-value pairs in its metadata so I’m not surprised its not automatically applied to the dataset.
So yeah, if you want to transform the patient, before or after segmentation you can use the vtkTransform class (http://www.vtk.org/doc/nightly/html/classvtkTransform.html) and set the translation/rotation/scale. You can find some sort-of-ok examples under http://www.vtk.org/doc/nightly/html/c2_vtk_e_8.html#c2_vtk_e_vtkTransform. Personally, I think I would try to segment the patient anatomy prior to transforming, especially since you’ll have to visualise the results which would be much tricker once the patient is in an arbitrary plane (and you’d have to use oblique planes to view 2D slices).
LikeLike
Hey There! Thanks for the great tutorials. Is there a way in SimpleITK to read Nifti files. I performed the segmentation using ITK and I have the ground truth in NIFTI format. I tried using nibabel but it always gives me memory error. I basically want to match the segmentation results with the groundtruth to get the Dice Score.
Appreciate the help!
Sidh
LikeLike
Hey Sidh, as far as I can see ITK does have a NIFTI class (https://itk.org/Doxygen/html/classitk_1_1NiftiImageIO.html) so its mostly a matter of whether this was ported to SimpleITK.
What sort of error do you get with NiBabel? Are you sure you have enough memory to read it in?
LikeLike
Hmm the NIFTI class looks neat. May be I should work with this rather than working with nibabel and SimpleITK. I get an out-of-memory error when loading image.
`img = nb.load(filename)
imgdata = img.get_data()`
OR
`imgdata = np.asarray(img.dataobj)`
The problem is I have the ground truth in nifti format. As i said I wanted to get the dice score, I was working with DICOM and sitk. I can change the DICOM into nifti, which would be an obvious thing to do, but the memory error is really irritating. I checked the memory on Task Manager (not the best way but it wouldn’t be that off the mark).
Additionally, do you know of a better way than the below code to change sitk image to vtk.
def itkImageToVtk(image):
arrayData = sitk.GetArrayFromImage(image)
print arrayData.flags
arrayDataC = np.array(arrayData[:,:,1], order=’C’)
print arrayDataC.flags
ArrayDicom = numpy_support.numpy_to_vtk(arrayDataC)
This is pretty dubious as I have to concat the arrays to make a 3D matrix. I really just want this to get the beautiful marching cube working.
Thanks a lot for the quick answers.
Much appreciated. You understand fellow BME people unlike those doctors 😉
LikeLike
Hey Sidh, thanks for the kind words. You mentioned in your other comment (https://pyscience.wordpress.com/2014/10/19/image-segmentation-with-python-and-simpleitk/comment-page-1/#comment-436) that you’re using 32bit Python. Any chance you’re going past the 2GB heap size? Chances are that the NumPy conversion copies the memory doubling the usage.
Alternatively you could simply use 3DSlicer to convert the Nifti file to another format such as MHD which can be easily read by SimpleITK.
I’ve committed the half-assed material I was putting together on the conversion between NumpySimpleITKVTK under https://bitbucket.org/somada141/pyscience/commits/95f7b8e31af8029d10ee16becc582b317eaf5557?at=master . Take a look on the conversion(s) there.
Lastly, if you don’t want to load each individual DICOM and concatenate the strings you could use VTK and `vtkDICOMImageReader` to read a whole directory of images in one go as shown under https://pyscience.wordpress.com/2014/09/08/dicom-in-python-importing-medical-image-data-into-numpy-with-pydicom-and-vtk/
Hope the above helps 🙂
LikeLike
Thank you for your reply. I am sure it is the heap problem because it comes at run time. I can load .nii.gz file using sitk but now I get a weird exit code which I can’t catch. Please look at this so question that I posted and let me know what you think.
http://stackoverflow.com/questions/39921855/unhandled-exit-code-when-loading-nifti-with-simpleitk-in-python?noredirect=1#comment67201356_39921855
LikeLike
Hi Adam
Thanks for post ,finally I am able to get image and label files for MRI data ..they both are in .mha format. Can you please suggest me some pre processing steps? And I have to do pre processing on both right label and image?
LikeLike
Hey Sagar, I’d probably just suggest the same pre-processing steps I noted on the post, i.e., smoothing/denoising to clear up outlier-pixels and weird island, followed by thresholding to isolate the anatomy from the background. Then as far as automatic segmentation goes thresholding, region-growing, and possibly kmeans-clustering are your best bets.
LikeLike
That is very interesting, You are an excessively
professional blogger. I’ve joined your feed and sit up for looking
for extra of your wonderful post. Also, I’ve shared your
web site in my social networks
LikeLike
Thanks a lot for the kind words 🙂 sadly I haven’t had the time to write in ages but you never know 🙂
LikeLike
Hey,My friend
First , thank you for your tutorial.
I have some questions and I want to get your help!
1, I am a novice in medical image segmentation, And I want to use SVM to segment MR brain tissue(White matter,Gray matter,Cerebrospinal fluid ). here i have some .mhd files including T1.mhd and T1_1mm.mhd,The SVM need to have feature vectors and labels for training, The Dataset description I have downloaded have ground truth.but i don’t konw what’s the meaning of it.
2, if i want to extract ground truth (labels) as each voxel’s label(am i right??), how can i do that.
Thanks you!
Best wishes!
Jeffery
LikeLike
Hi Jeffery, I’m not sure what the issue is. If the T1 data is pre-segmented then the labels should be stored in the .mhd files and reading those files would directly yield the labels. If you have a link to such an MHD file I could take a look.
LikeLike
can you pl tell me how to convert .raw file image data to dicom series with .mhd?
LikeLike
You mean you have an .mhd file with the accompanying .raw file? You can convert them to DICOM using an approach like this: https://stackoverflow.com/questions/14350675/create-pydicom-file-from-numpy-array
LikeLike
hi. i have .raw file with .mhd from VESSEL12.grandchallenge.org site. .raw file images are not numpy array i suppose, not sure, pl suggest
LikeLike
one more thing sir, what to install to make import gui work?
LikeLike
erm not sure what you mean but I assume you’re referring to the VTK interactive window. Nothing to import apart from VTK but you’d have to change the way the rendering is invoked. You can find an example on how to do it in the last cell under this notebook: http://nbviewer.jupyter.org/urls/bitbucket.org/somada141/pyscience/raw/master/20140917_RayTracing/Material/PythonRayTracingEarthSun.ipynb
LikeLike
i followed your tutorials and trying lung volume segmentation. but during region growing trachea and broncholi (smaller components) are also present. pl tell me how to remove smaller areas. i think connected comp of simpleitk has to be used but don’t know how to implement the code. kindly guide
LikeLike
I followed your tutorial but simpleitk is not reading my .mhd file. my .mhd file is with .zraw file which i got from MITK software. Can you please tell me difference between zraw and raw.
LikeLike
Hi Hitesh, `.zraw` files are compressed `.raw` files. Looking into whether SimpleITK supports these.
LikeLike
Can’t find any info on whether zraw is supported by ITK/SimpleITK but it might well work. Alternatively you could try and use VTK to read them which seems to support them.
LikeLike
Thank you
LikeLike
Pingback: Image Segmentation, Registration and Analysis – Mi(Medical imaging) Favourite Things
Hi,
Thank you for the post.
Can you tell me how to read multiple images in SimpleITK because I want to do multiple image registration or in simple words how can I register multiple moving images with a single fixed image or is it possible to register a moving image with already registered image considering it as a fixed image in SimpleITK?
LikeLike
Hi Akshay, I haven’t actually done any image-registration with SimpleITK/ITK before. There are some good tutorials and guides under https://simpleitk.readthedocs.io/en/master/Documentation/docs/source/registrationOverview.html#lbl-registration-overview but I haven’t tried it. I’ve used Elastix (http://elastix.isi.uu.nl/) before with good results but it may not be what you’re looking for.
LikeLike
Thank you for your reply
LikeLike
Hi Thanks a ton for all your post. Your post is helping me understand the basics of segmentation especially for medical images. When it comes to 3D image segmentation how can we extract features and calculate measures from the segmenting 3D images. Are there any specific examples or guidelines around it?
LikeLike
It depends on what you mean by ‘calculate measures’. 3D segmentation isn’t all that different from 3D but do you mean statistics like volume, area, weight, number of voxels? I think you need to be a little more specific 🙂
LikeLiked by 1 person
Thanks for your reply. Sorry for not being specific. In terms of post processing I wanted to know if I need to iterate over each 2D slices to extract measures like Area (ROI) in each slices, intensities, textures etc. Does SimpleITK has some API to collect all such stats at 3D image or we need to iterate over each 2D slices?
LikeLike
SimpleITK definitely has image statistics filters that should work both in a 2D and a 3D label-field. Eg check the process under http://insightsoftwareconsortium.github.io/SimpleITK-Notebooks/Python_html/03_Image_Details.html. You can check the different statistics filters under https://simpleitk.readthedocs.io/en/master/Documentation/docs/source/filters.html
LikeLiked by 1 person
This is really cool and helpful. Thanks a ton! Yes, I can see there are api where I can extract info in both label field. One thing I could not figure out is when I apply segmentation on entire 3D image, post segmentation I want to know on which slices of 3D image the label has been applied. As an example: in 50 slices image segmentation label are only between , let say 20-25, How can I extract such information post segmentation?
LikeLike
Hm once you’re working on a label field then what you’re describing above sounds like extracting the bounding-box out of the label. It might be possible with LabelShapeStatisticsImageFilter but I haven’t managed to find much info on it.
Alternatively I would convert the image to NumPy, threshold it to just the label you’re interested though something like `img_nump[img_nump != 1] = 0` and then find the minimum and maximum index across each axis you’re interested in (through numpy.argmin and numpy.argmax).
LikeLiked by 1 person
Perfect, yeah I can try capture bounding box info that will give me range of index across each axes. Your suggestion using numpy is excellent too. Will explore in that direction! Thanks much!!
LikeLike
Hi I found that LabelShapeStatisticsImageFilter gives that information. If you use GetBoundingBox method on shape_stats data we get from the filter we get info as [xstart, ystart, start, xsize, ysize, zsize]. So from start -> z Size we know depth of images that are labelled.
Reference: https://github.com/SimpleITK/SimpleITK/issues/606
LikeLike
Oh that’s awesome thanks for sharing that!
LikeLike
You’re welcome! Btw I bumped into another issue, thought to share if you have come across this, I used basic thresholding simple ITK api providing upper, lower and outside values to segment 3D image. It gave me thresholded image but the labels were all in the lower and upper range. I need the labelling to be constant for all pixel intensities between upper and lower. To achieve that I tried using Aggregate Label filter but came to know from API docs, that it does not work in signed 16 bit pixel type. So to bypass this I am thinking to read this thresholded image into simpleITK array or. numpy array. Iterate over this array and give pixel values, that are within the range with label number I wish and then again create simpleITK image out of it this new array. Hope I am going in right direction 🙂
LikeLike
Nevermind! I used BinaryThresholding 😉
LikeLike
Sorry Jiten was at work, glad you figured it out 🙂
LikeLike
Hi there. thank you very much for your helpful explanation of the various systems for 3D images. I need help from you, please. I have downloaded .mhd and .raw files from the internet. But I can’t load and preprocess all these images on my computer. I don’t have a CPU, I simply want to see some of the images and use for my final year thesis on lung cancer classification. anyone who can help, please.
LikeLike
Hi Tekle, I’m not sure why can’t you see the images on the machine you downloaded them on, and I definitely don’t get what you mean by “I don’t have a CPU”, but if you simply can’t use a downloadable viewer like Radiant (https://www.radiantviewer.com/) or Paraview (https://www.paraview.org/) then you can view DICOM images on your browser with online viewers like Imaios (https://www.imaios.com/en/Imaios-Dicom-Viewer) or DICOMViewer (http://dicomviewer.booogle.net/) or you can find others like them (some of which also support MHD) under https://medevel.com/14-best-browser-web-based-dicom-viewers-projects/.
Hope this helps,
Adam
LikeLiked by 1 person
Thank you very much for your immediate response sir. It was to say I don’t have GPU. I am trying to load from my directory and preprocess the .mhd and .raw image using the python library on jupyter notebook. I downloaded those images from the luna16 lung cancer dataset repository. it is not a DICOM file sir. But unfortunately, I am getting an error on my python code when I try to run it. let me share the python code if you can observe and identify the possible problem.
import os
import numpy
import SimpleITK
import matplotlib.pyplot as plt
%pylab inline
”’
This funciton reads a ‘.mhd’ file using SimpleITK and return the image array, origin and spacing of the image.
”’
def load_itk(“C:/Users/tek/SampleNodule”):
# Reads the image using SimpleITK
itkimage = sitk.ReadImage(“C:/Users/tek/SampleNodule”)
# Convert the image to a numpy array first and then shuffle the dimensions to get axis in the order z,y,x
ct_scan = sitk.GetArrayFromImage(itkimage)
# Read the origin of the ct_scan, will be used to convert the coordinates from world to voxel and vice versa.
origin = np.array(list(reversed(itkimage.GetOrigin())))
# Read the spacing along each dimension
spacing = np.array(list(reversed(itkimage.GetSpacing())))
return ct_scan, origin, spacing
### Here is the error that is popped up when i run this code.
File “”, line 10
def load_itk(“C:/Users/tek/SampleNodule”):
^
SyntaxError: invalid syntax
LikeLike
hi.
I am a beginner in radiomics and images. And currently i’m working on segmentation of gallbladder from ct scan images. can you please help me with how to procees? it would be of great help. thank you.
LikeLike